Spring 2025 Update: AI Is Cracking “Inscrutable” Proteins — Here’s What People Still Do Best
New AI systems are getting better at describing previously indecipherable proteins. We pair those predictions with human labeling to convert patterns into testable function.
Shawnak Shivakumar
4/3/20251 min read
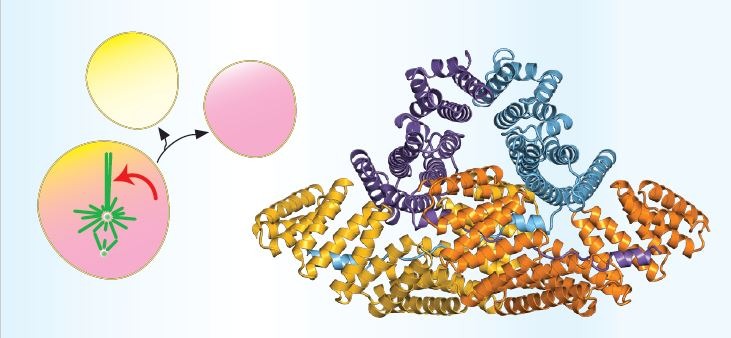
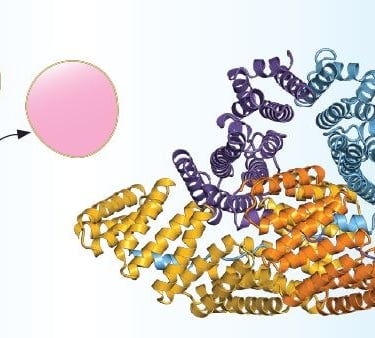
This spring brought a wave of encouraging coverage: AI tools are getting sharper at detecting and describing previously undiscovered proteins and their features. That’s great news for anyone who’s stared at a low-confidence model and thought, “So what?” We’re moving from “folds exist” to “features matter.”
But there’s a gap between pattern and purpose. On one side are interaction-savvy predictors like AlphaFold 3, which model complexes with proteins, nucleic acids, ions, and small molecules. On the other are real-world labels — the pocket boundaries, catalytic residues, and surface chemistry that make a prediction useful to an experimentalist. Our community sits in that gap. We take proposed sites and stress-test them: is the geometry compatible with a ligand family? Is a cavity hydrophobic in the right places? Does the residue micro-environment agree with the reaction chemistry?
Practically, that means feeding human-curated positives/negatives back into training and ranking. It also means triaging long-tail proteins where homology is thin and confidence is lumpy. The result is simple: more testable hypotheses per week. AI is widening the searchlight; people are telling it where to linger.
Contact
shawnak@darkprotein.org
shawnak.shivakumar@gmail.com
