Finding druggable pockets at scale — smarter attention, better sites, faster discovery
Finding druggable pockets at scale — smarter attention, better sites, faster discovery
Shawnak Shivakumar
2/12/20251 min read
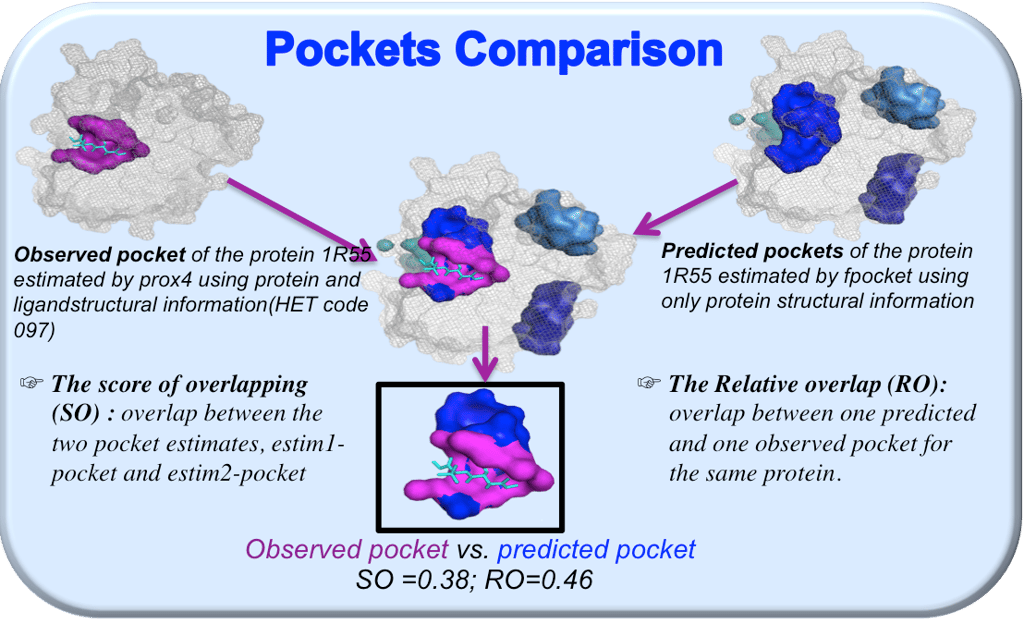
A wave of new work has pushed binding-site prediction beyond heuristics. One example: a 2024 study introduced pocket-guided explicit attention, boosting accuracy by steering models toward ligandable cavities. That theme continues into 2025 with fresh methods aimed at binding-site-agnostic docking and pocket identification: exactly the space where open access research excels.
Here’s how we plug in. Our Level-1 tasks focus on known proteins/known sites (teaching the intuition: where do real ligands sit, what residues shape the chemistry?). Level-2/3 tasks then move into uncertain pockets from newer predictors. Human reviewers flag false positives (e.g., clefts with no chemistry) and up-vote cryptic sites that only open in certain conformations. Those judgments become labels that help pocket models generalize: especially on dark proteins with few homologs.
The payoff is practical: better pockets shorten the path from “mystery protein” to testable small-molecule hypotheses. That means faster iteration for neglected targets and organisms that rarely get industry attention.
Contact
shawnak@darkprotein.org
shawnak.shivakumar@gmail.com
